Virus structure and cell entry
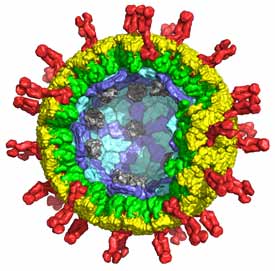
Virus particles carry genetic material from one cell to another. The key structural distinction is between viruses that have lipid-bilayer membranes and those that lack them –“enveloped” and “non-enveloped”, respectively. These differences reflect different mechanisms of cell entry and different pathways of assembly and maturation. Enveloped viruses enter by membrane fusion, either from an internal compartment following an endocytic step, or at the cell surface. Non-enveloped viruses require some form of membrane “perforation”. For these viruses to penetrate a cell, a large macromolecular complex (either a subviral particle or just the viral genome) must cross a cellular membrane to access the cytosol, and some mode of local membrane disruption must accompany the translocation. We study the mechanisms of membrane fusion and perforation, directing our attention currently to flaviviruses (especially dengue and West Nile viruses) and double-strand RNA viruses (rotavirus, in particular).
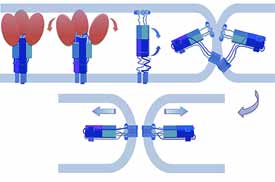
Viral membrane fusion
Membrane fusion is thermodynamically favorable, but it generally presents a high kinetic barrier. Fusion proteins lower this barrier and thus are catalysts for the merger of two bilayers — “suicide” catalysts in the case of viral fusion proteins. Sensitive measurements of fusion kinetics, like careful analysis of enzyme kinetics, can yield information about mechanism, by examining effects of directed mutations on rates, cooperativity, and so forth. We have devised a single-virus-particle fusion assay to study fusion in this way and applied it to influenza virus (Floyd et al, 2008; Ivanovic et al, 2013 and 2015) and to West Nile and dengue viruses (Chao et al, 2014 and 2018). Our on-going work is an effort to link cryo-EM structures of flavivirus envelope protein membrane-interacting regions with steps in the fusion pathway and ultimately to understand more thoroughly the mechanism of fusion inhibition by small-molecule inhibitors (Schmidt et al, 2012; Chao et al, 2018).
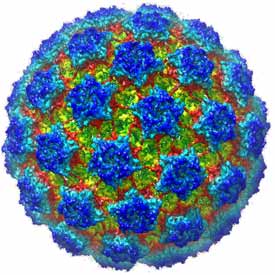
Non-enveloped virus entry
A principal objective of our research on virus structure is a molecular description of the earliest events leading to infection of a cell: attachment, uptake, and penetration into the cytosol. Rotavirus, a major cause of infectious infant diarrhea, is a particularly suitable non-enveloped virus for deriving such a “molecular movie”. We have used electron cryomicroscopy (with Nikolaus Grigorieff, Brandeis) to obtain an atomic model for the complete rotavirus particle, taking advantage of a number of high-resolution structures (from x-ray crystallography) of individual structural proteins and protein fragments and of the intact inner capsid particle. The structural analysis has enabled us to design experiments using live-cell imaging (with Tomas Kirchhausen, Children’s Hospital: Abdelhakim et al, 2014; Salgado et al, 2017 and 2018) to follow molecular events during virus uptake and penetration into cells in culture. Our on-going work is an effort to connect conformational changes in components of the virus particle, as defined by structures from cryo-EM and x-ray crystallography, with stages of entry as defined by live-cell imaging.
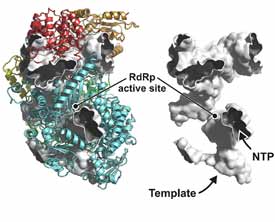
Viral RNA polymerases
Structures of viral RNA polymerases define aspects of transcription and replication that differ from related events in the host cell. We are expanding our study of the polymerases of negative-sense, single-segment, RNA viruses (a collaboration with Sean Whelan, Harvard Medical School: Liang et al, 2015) to work out a full picture of the stages of transcription and replication and of the interaction of the polymerase with the template RNP. We are also studying structural events within the virion linked to the polymerization cycle of the rotavirus polymerase (for our earlier work, see Estrozi et al, 2013